Aitomistic Updates: IRC Calculations, AIQM3, and New ML Research
Here is your weekly dose of exciting updates around aitomistic simulations from our team, including IRC, a video lecture and new publication, and preview of upcoming features.
IRC (intrinsic reaction coordinate) calculations
Many of you use Aitomistic for performing transition state calculations with AIQM2 and other methods. The next logical step is performing IRC calculations on the obtained structure – something that was unavailable for online computations until recently. We have listened to your requests, and we are happy to share that this essential feature is now available for online computations and in open-source MLatom, implemented by Xinxin – the author of AIQM2!
IRC calculations are as simple as a three-line input file or a couple of lines in a Python script, with rich features for plotting and analysis. A beautiful tutorial is available online.
Improvements on the Hub & Lab
We are continuously improving Hub & Lab to make the user experience smoother. Here, your feedback is crucial - if you do not tell us that something is not working for you, we will not be able to make it work. Recently, we have substantially simplified operations with files and folders so that you can now easier manage them, e.g., copy and paste, duplicate, rename, etc, with as little hassle as possible.
Release of Aitomic add-ons to MLatom, version 0.92
We have also upgraded add-ons to MLatom, which now contain new AIQM3 method. The new method retains the performance of AIQM2 but is extended to more elements, now covering CHNOFSCl and targeting coupled-cluster accuracy. Upgrade includes many further improvements such as support of higher versions of PyTorch.
We have also updated its installation instructions at https://aitomistic.com/mlatom/addons.html.
New publication on ML-NACs
On Monday, a paper describing a novel methodology for machine learning of nonadiabatic couplings with high accuracy based on physics-informed descriptors (energy gradient difference) appeared in JPCL. Methodology is based on implementations in MLatom, including FSSH (fewest switches surface hopping), which can be performed entirely with ML, with QM, and in hybrid ML/QM combinations as implemented by Jakub (paper’s first author).

Read more in the blog and in the paper:
Jakub Martinka, Lina Zhang, Yi-Fan Hou, Mikołaj Martyka, Jiří Pittner*, Mario Barbatti*, Pavlo O. Dral*. A Descriptor Is All You Need: Accurate Machine Learning of Nonadiabatic Coupling Vectors. J. Phys. Chem. Lett. 2025, 16, 11732–11744. DOI: 10.1021/acs.jpclett.5c02810.
Online lecture on MD with ML
If you wish to learn more about MD (molecular dynamics) with ML, which you can do on Aitomistic Hub & Lab, we have given an online lecture. The lecture covered many important topics, such as pitfalls and state-of-the-art of machine learning interatomic potentials including the universal models and their fine-tuning. In addition, the lecture has covered emerging, radically different ways of performing and analyzing molecular dynamics by directly learning molecules in four-dimensional spacetime.
Did you know that we have official accounts on several video platforms as well as Prof Pavlo Dral's account? You can subscribe to watch more videos.
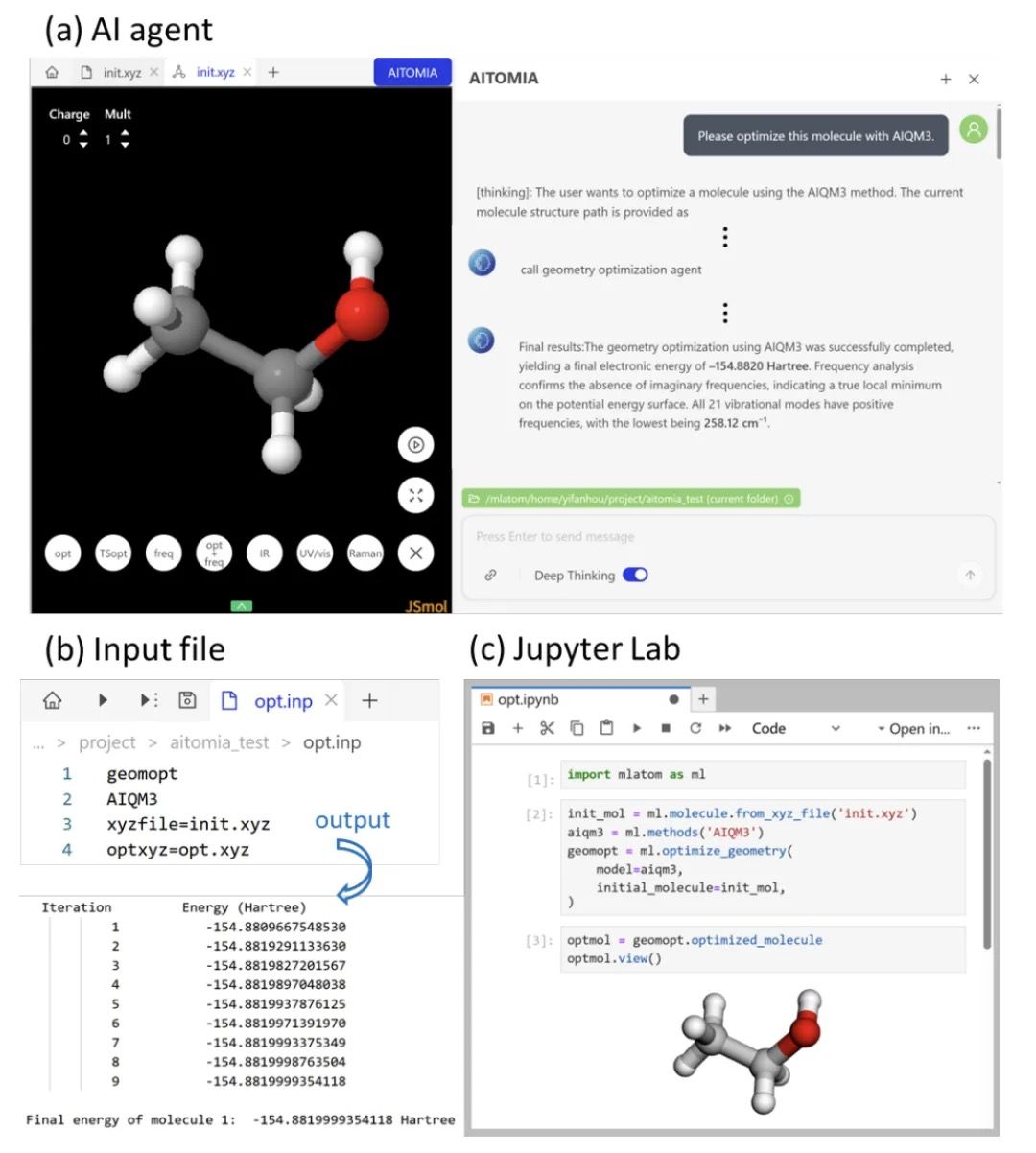
Preview of upcoming features
In the coming days and weeks, we will roll out several big upgrades such as improved AI agents, optimization of conical intersections, upgraded OMNI-P2x models for excited-state simulations with tutorials about UV/vis spectra simulations and fine-tuning for NAMD, etc. Stay tuned!
Please also let us know what you would like us to improve to make your experiences with Aitomistic better. Simply reply to this email or join us on Slack to directly chat with us.